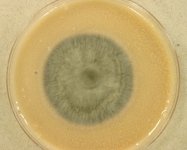
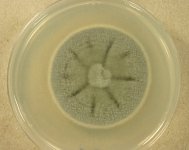
Search Details
| UAMH Number: | 11713 |
|---|---|
| Species Name: | Acephala applanata |
| Type: | |
| Synonyms: | |
| Taxonomy: | FUNGI Ascomycota, Leotiomycetes, Helotiales |
| Strain History: | Sieber, T. (T1_52_2) -> UAMH |
| Substrate: | roots of Picea abies (Norway spruce) | Location: | SWITZERLAND Bodmerenwald (GEO: 46.972,8.835) |
| Isolator: | |
| Isolation Date: | |
| Date Received: | 2013-04-09 |
| Characters: | MOLECULAR SYSTEMATICS Population genetic analysis of the Phialocephala fortinii complex - Gruenig CR, Duo A, Sieber TN, Fung Gen Biol 43:410-421, 2006 // MOLECULAR SYSTEMATICS Population genetic analysis of the Phialocephala fortinii complex - Queloz V, Duo A, Gruenig CR, Molecular Ecology Resources 8:1322-1325, 2008 // MOLECULAR SYSTEMATICS Species recognition within Phialocephala fortinii complex - Gruenig CR, Brunner PC, Duo A, Sieber TN, Fungal Gen Biol 44:773-788, 2007 // MYCORRHIZAE effects of P. subalpina and other PAC on survival and performance of Norway spruce seedlings - Tellenbach C, Grunig CR, et al, Environmental Microbiology 13(9): 2508-2517, 2011 (Click for publications citing UAMH 11713) |
| Compounds: | |
| Cross Reference: | |
| Collections: | Living Strains; Dried Herbarium Material |
| Pathogenic Potential: | Human: no | Animal: no | Plant: no |
| Biosafety Risk Group: | RG1 (check the PHAC ePATHogen Risk Group Database for updates) |
| Regulatory Requirements: | Canadian requesters must provide PHAC Pathogen and Toxin License Number (see: https://www.canada.ca/en/public-health/services/laboratory-biosafety-biosecurity/licensing-program.html) prior to shipment. International requesters must provide all legally required importation documentation prior to shipment. Plant pathogenicity status may be verified by using the USDA Agricultural Research Service (ARS) Fungal Database |
| MycoBank ID: | 344358 |
| Sequences: | >UAMH11713_DQ274839_bTub ACATCTGCATGAGAACCCTCAAGCTCTCCAACCCCTCTTACGGAGATCTCAACCACTTGGTCTCTGCCGTCATGTCGGGTGTCACTACCTGTCTCCGTTTCCCAGGTCAACTCAACTCTGATCTCCGAAAGTTGGCCGTCAACATGGTTCCCTTCCCTCGTCTCCATTTCTTCATGGTTGGCTTCGCTCCTCTTACCAGCCGTGGTGCTCACTCTTTCCGTGCCGTCACGGTCCCAGAGTTGACCCAGCAAATGTATGACCCAAAGAACATGATGGCTGCGTCCGATTTCCGTAACGGTCGCTACTTGACTTGCTCTGCTATCTTGTAAGTCTTCATAGCTCATTTTTGATGCCGCATGCTAACTTCTTCCAGCCGTGGTAAGGTCTCCATGAAGGAGGTTGAGGACCAAATGCGCAATGTTCAGAACAAGAACTCCTCCTACTTCGTCGAGTGGATTCCCAACAACGTCCAAACCGCACTCTGCTCCATTCCACCTCGTGGCCTCAAGATGTCTTCCACTTTCGTCGGAAACTCAACCTCCATCCAAGAGCTCTTCAAGCGTGTTGGTGACCAGTTCACTGCC >UAMH11713_DQ274790_bTub ACATCTGCATGAGAACTCTCAAGCTCTCCAACCCCTCTTACGGAGATCTCAACCACTTGGTCTCTGCCGTCATGTCGGGTGTCACTACCTGTCTCCGTTTCCCAGGTCAACTCAACTCTGATCTCCGAAAGTTGGCCGTCAACATGGTTCCCTTCCCTCGTCTCCATTTCTTCATGGTTGGCTTCGCTCCTCTTACCAGCCGTGGTGCTCACTCTTTCCGTGCCGTCACGGTTCCAGAGTTGACCCAGCAAATGTATGACCCAAAGAACATGATGGCTGCCTCCGATTTCCGTAACGGTCGCTACCTGACTTGCTCTGCTATCTTGTAAGTCTTCATAGCTCATTTTTGATGCCGCATGCTAACTTCTTCCAGCCGTGGTAAGGTCTCCATGAAGGAGGTTGAGGACCAAATGCGCAATGTTCAGAACAAGAACTCCTCCTACTTCGTCGAGTGGATTCCCAACAACGTCCAAACCGCGCTCTGCTCCATTCCACCTCGTGGCCTCAAGATGTCTTCCACATTCGTCGGAAACTCAACCTCCATCCAAGAGCTCTTCAAGCGTGTTGGTGACCAGTTCACTGCC >UAMH11713_DQ274573_EF1a CTCGGTGGGGTAAAATCCTTCGCCCGGGCCATCGTCACGACAACATCATGATTGATGCCCAACGCACATACCCTTTCACAACATCCATACCTTTTCACAACATACATTGTCTTTCGATACAATGCTGACCCTTTCAATAGGAAGCCGCCGAGTTGGGCAAGGGTTCTTTCAAGTACGCATGGGTTCTTGACAAGCTAAAGGCCGAGCGTGAGCGTGGTATCACCATCGATATTGCTCTCTGGAAGTTCGAGACCCCAAAGTACTATGTCACCGTCATTGGTATGTCTTCGCAGATGCATTGCATCATCTTCATTGCTAACTCCTTGCAGATGCTCCCGGTCACCGTGATTTCATCAAGAACATGATCACTGGTACTTCCCAGGCCGATTGCGCTATTCTCATCATCGCTGCCGGTACTGGTGAGTTCGAGGCTGGTATCTCCAAGGATGGCCAGACTCGTGAGCACGCCCTCCTCGCCTACACCCTTGGTGTCAAGCAACTTATCGTCGCCATCAACAAGATGGACACCACCAAGTGGTCCGAGGCTCGTTACCAAGAAATC >UAMH11713_DQ274764_pPF-076 CCAACGCTTCGGCAGGACTGCCCATTGAATATTTCTCGCCAGAGTTGAAAGGAGTTTCCTCGGAGTCGAGATCGAGTGAAGATTGTACTGGAAGGCCGGGCGTTGTGCTGGGATCGTTGTTATTGCGCGAGGACTGTTGCTGTTTCATTCGAAGATAGACGTGAAAATGGATGTGAGAAATGGATGCAATCAAAGGTGCTGGTATGTAGGTAACGAAAGCCTTGAGTCCGGAAAGGCTAGCATCGTCTGCACCAGTCCCATTCCCCACAATGAAGGTATTGTACTCGAAGAAGATGCGGATAGCTTCGCTGTGAATCGCTTTGTTGACGTACAGGATGCTCCCTCTGGCATCTGAGAGGAGAAGCTCCTTGAAAGATCTGCGTCGTGGAGTCGCAATCAGTATCGGTTCTTTGTGGACGAGGAGAAGTCGATAAATTTGACTCCGTATTTCTGGCGGCACAGCCATCAGCACTCCTCCT |
IMAGES: