Search Details
| UAMH Number: | 6715 |
|---|---|
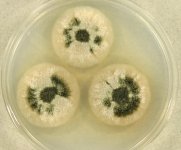
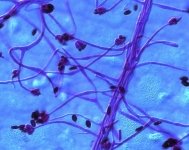
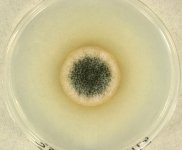
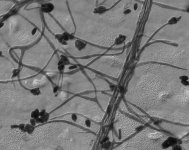
| Species Name: | Stachybotrys chartarum |
| Type: | |
| Synonyms: | Memnonium sphaerospermum / Oidium chartarum / Oospora chartarum / Sporocybe lobulata / Stachybotrys alternans / Stachybotrys alternans var. alternans / Stachybotrys atra / Stachybotrys atra var. atra / Stachybotrys atra var. brevicaule / Stachybotrys atra var. brevicaulis / Stachybotrys atrogrisea / Stachybotrys atrogriseus / Stachybotrys atrus / Stachybotrys atrus var. atrus / Stachybotrys atrus var. brevicaulis / Stachybotrys lobulata / Stachybotrys lobulata var. angustispora / Stachybotrys lobulata var. lobulata / Stachybotrys lobulata var. macra / Stachybotrys lobulatus / Stachybotrys lobulatus var. angustisporus / Stachybotrys lobulatus var. lobulatus / Stachybotrys lobulatus var. macrus / Stilbospora chartarum / Synsporium biguttatum / Torula chartarum / Trichosporum sphaerospermum |
| Taxonomy: | FUNGI Ascomycota, Sordariomycetes, Hypocreales, Stachybotryaceae |
| Strain History: | D. Inglis (2J2) -> Poznanski, S. -> UAMH |
| Substrate: | RCS strip, indoor air maintenance crawl space | Location: | CANADA Saskatchewan, Saskatoon (GEO: 52.133,-106.67) |
| Isolator: | D. Inglis (2J2) |
| Isolation Date: | 1991-02-19 |
| Date Received: | 1991-03-15 |
| Characters: | APPLICATION Cited in patent US20120129718 Qualitative/Quantitative Detection of Fungal Species - A. Cywinska // BIODIVERSITY indoor air fungi - // MOLECULAR SYSTEMATICS detection and phylogeny of Stachybotrys - Haugland & Heckman, Molecular & Cellular Probes 12:387-396, 1998 // MOLECULAR SYSTEMATICS hemolysis, toxicity & RAPD analysis - Vesper et al., Appl. Env. Microbiol. 65:3175-3181, 1999 // MOLECULAR SYSTEMATICS PCR assay for specific detection - Black JA, Dean TR, Foarde K, Menetrez M, Mycol Res 112:845-51, 2008 // MOLECULAR SYSTEMATICS phylogeny of Stachybotrys chartarum - Koster, Scott, Wong et al, Can. J. Bot. 81:633-643, 2003 // MOLECULAR SYSTEMATICS UAMH 6715, 7900, 7568, 7598 demonstrate similar sequence in ITS2 region and differ by one bp from ATCC 9182 - Haugland & Heckman, Molecular & Cellular Probes 12:387-396, 1998 // PIGMENT black - (Click for publications citing UAMH 6715) |
| Compounds: | |
| Cross Reference: | EPA 280 // NC-2 |
| Collections: | Living Strains; Dried Herbarium Material |
| Pathogenic Potential: | Human: no | Animal: no | Plant: no |
| Biosafety Risk Group: | RG1 (check the PHAC ePATHogen Risk Group Database for updates) |
| Regulatory Requirements: | Canadian requesters must provide PHAC Pathogen and Toxin License Number (see: https://www.canada.ca/en/public-health/services/laboratory-biosafety-biosecurity/licensing-program.html) prior to shipment. International requesters must provide all legally required importation documentation prior to shipment. Plant pathogenicity status may be verified by using the USDA Agricultural Research Service (ARS) Fungal Database |
| MycoBank ID: | 306362 |
| Sequences: | >UAMH06715_AY180249_bTub ATCTCTGGCGAGCACGGCCTCGACAGCAATGGTGTCTACAACGGTACTTCCGAGCTCCAGCTCGAGCGCATGAGCGTGTACTTCAACGAGGTATGTGAAATGGATTGAGGCAACTGCCTGATTGCACCTCTCGGCTCACAATTTCCCCAGGCTTCCGGCAACAAGTATGTCCCTCGCGCCGTCCTTGTCGATCTCGAGCCCGGTACCATGGACGCCGTCCGGGCTGGCCCATTCGGTCAGCTCTTTCGACCCGACAACTTCGTCTTCGGCCAGTCTGGTGCTGGCAACAACTGG >UAMH06715_AY180257_CAM AGGTCTATCTCAATAGCTAAGCTTACCTCTAGCTAAGGCTAACGCGCTGCCACAGGACAAGGATGGTGATGGTAAGTGGTGAAGAGCCATGCGTCGAACGTCATTAGTCCTAGATCAACATTTCTCTCGATCTCTTCCTCGACCGCTCTCCTCTAGTCACTCCTTTCGCCTCTCGACATTATACGTTTCGGGAAAAATCAGCTGATCCATGATTCAACAGGTCAGATTACCACCAAGGAGCTGGGCACTGTCATGCGCTCCCTAGGCCAGAACCCCTCCGAGTCTGAGCTCCAGGACATGATCAACGAGGTTGATGCCGACAACAACGGCACCATCGACTTTCCTGGTAGGTTACTATTACCAGTGTCGAAAGGCTACGAGTCGACGGCTTTTGTCATTCATTTTATCAATTGCTGTACTAACCCGTATCTATAGAATTCCTCACCATGATGGCGCGCAAG >UAMH06715_AY180265_EF1a ACGTCGACTCCGGCAAGTCGACCACCACTGGTCACTTGATCTACCAGTGCGGTGGTATCGACAAGCGTACCATCGAGAAGTTCGAGAAGGTAAGCTCCTCTTGTTTCCCCTGGGCCCCCTTCACTCGAGGCGCGCACTTTTTGCCCCGCTTCCTGGACAAAAATTTCCCCCTCCTCTCGTGGTGTCTCATCTGCCTGCGGGGCACTTTACCCCGCCTGCTCTTTGGAGCATGAGCACAGCAAAGCACTTGCCTCACATTAACTCGGTGTTCCACCGTCAAAAGCACTGTATTTGTTACTGACTCGCATGACAGGAAGCTGCCGAACTCGGCAAGGGTTCCTTCAAGTACGCCTGGGTGCTTGACAAGCTCAAGGCTGAGCGCGAGCGTGGTATCACCATTGATATTGCTCTCTGGAAGTTCGAGACTCCCAAGTACTATGTCACCGTCATTGGTATGTTCCAACCAACATGCCTCATCATCGCTTCGATCCTAACAATTTGCAGATGCCCCTGGTCACCGTGACTTCATCAAGAACATG >UAMH06715_AY176729_LSU GAAACCAACAGGGATTGCCCCAGTAACGGCGAGTGAAGCGGCAAAAGCTCAAATTTGAAATCTGGCTCCTTTAGGGGTCCGAGTTGTAATTTGCAGAGGATGCTTCGGGAGCGGCCACGGTTTAAGTGCCTTGGAACAGGACGTCATAGAGGGTGAGAATCCCGTCTTGAATCGTTGGTCCGCGCCCATGTGAAGCTCCTTCGACGAGTCGAGTTGTTTGGGAATGCAGCTCTAAGCGGGTGGTAAATTTCATCTAAAGCTAAATATTGGCCGGAGACCGATAGCGCACAAGTAGAGTGATCGAAAGGTTAAAAGCACCTTGAAAAGGGAGTTAAACAGCACGTGAAATTGTTGAAAGGGAAGCGCTTGCGGTCAGACTCGGGTACAGGGTTCAGCGGGTGCGTGTCACCCGTGTACTCCCTGTTTCTCGGGCCAGCATCAGTTCTGACGGCCGGTCAAAGGCCTCTGGAATGTGTCGTCTCTCGGGACGTCTTATAGCCAGGGGTGCAATGCGGCCTGTTGGGACTGAGGAACGCGCTTCGGCTCGGATGCTGGCATAATGGCCGTAAGCGGCCCGTCTTGAAACACGGACCAAGGAGTCTAACATCCACGCGAGTGTTCGGGTGTCAAACCCGTGCGCGAAGTGAAAGCGAACGGAGGTGGGAGCCTTAGGGCGCACCATCGACCGATCCTGAAGTCTTCGGATGGATTTGAGTAGGAGCGTGGCCGTTGGGACCCGAAAGATGGTGAACTATGCCTGAATAGGGTGAAGCCAGAGGAAACTCTGGTGGAGGCTCGCAGCGGTTCTGACGTGCAAATCGATCGTCAAATTTGGGCATAGGGGCGAAAGACTAATCGAACCATCTAGTAGCTGGTTCCTGCCGAAGTTTCCCTCAGGATAGCAATAACGAAGTTTCCAGTTTTATGAGGTAAAGCGAATGATTA >UAMH06715_AY180260_SSU-LSU CTCCGTTGGTGAACCAGCGGAGGGATCATTACCGAGTTTACAACTCCCAAACCCTTATGTGAACCGTACCTATCGTTGCTTCGGCGGGAACGCCCCGGCGCCCTGCGCCCGGATCCAGGCGCCCGCCGGAGACCCCAAACTCTTGTGTTTTTTTCAGTATTCTCTGAGTGGCAAACGCAAAAATAAATCAAAACTTTTAACAACGGATCTCTTGGCTCTGGCATCGATGAAGAACGCAGCGAAATGCGATAAGTAATGTGAATTGCAGAATTCAGTGAATCATCGAATCTTTGAACGCACATTGCGCCCGTTAGCATTCTAGCGGGCATGCCTGTCCGAGCGTCATTTCAACCCTCAGGGTCCCCGTTCCGGCGGGGAACCTGGTGTTGGGGATCGGCCCGCCCCGCGCGGCGCCGTCCCCCAAATTCAGTGGCGGTCTCGCTGCAGCCTCCCCTGCGTAGTAGTTACAACCTCGCATCGGAGCTCAGCGCGGCCACGCCGTAAAACCCCCGACTTTCTGAACGTTGACCTCGGATCAGGTAGGAA >UAMH06715_AY180275_tri5 TTGATGATCTTCAGAGTGGCAACCCGCAAAAGCATCCCTGGTGGATGCTGGTCAACGAGCACTTCCCTAATGTGCTGAGACACTTTGGCCCTTTCTGCTCGCTAAACCTCATTCGCAGCACACTGGACTGTAAGTCTATACTCGACAATAGTCCCCCAAAATATTCCAAGAAAGAGCAAGAAAACTAACACGCTTCCACAATAGTCTTTGAAGGTTGCTGGATTGAGCAATATAACTTCCATGGCTTCCCAGGCTCCTTTGACTATCCTGGATTCCTTCGTCGTATGAATGGACTAGGACACTGTGTCGGAGGATCTTTGTGGCCAAAGGAAAACTTCAACGAGCAGGAGCATTTCTTGGAAATCACCAGCGCCATCGCCCAAATGGAAAACTGGATGGTTTGGGTTAA |
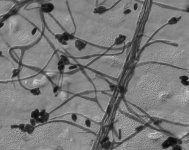
IMAGES: