<< back to search
IMAGES:
Search Details
add to cart
| UAMH Number: | 7863 |
|---|---|
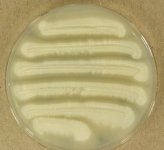
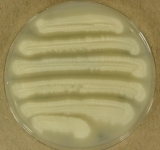
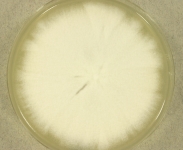
| Species Name: | Geotrichum candidum |
| Type: | |
| Synonyms: | Acrosporium candidum / Alysidium lactis / Botrytis geotricha / Dipodascus geotrichum / Endomyces geotrichum / Endomyces lactis / Endomyces lactis var. matalensis / Galactomyces candidum / Galactomyces candidus / Galactomyces geotrichum / Geotrichum asteroides / Geotrichum bryndzae / Geotrichum javanense / Geotrichum malti-juniperini / Geotrichum matalense / Geotrichum matalense var. chapmanii / Geotrichum novakii / Geotrichum redaelli / Geotrichum redaellii / Geotrichum silvicola / Geotrichum versiforme / Monilia asteroides / Mycoderma asteroides / Mycoderma malti-juniperini / Mycoderma matalense / Oidium asteroides / Oidium humi / Oidium lactis / Oidium lactis var. luxurians / Oidium matalense / Oidium nubilum / Oidium obtusum / Oidium suaveolens var. minutum / Oospora fragrans var. minuta / Oospora humi / Oospora lactis / Oospora lactis var. exuberans / Oospora lactis var. obtusa / Oospora lactis var. parasitica / Oospora matalensis / Oospora nubilum / Oosporoidea lactis / Pseudomonilia matalensis / Pseudomycoderma matalense / Scytalidium candidum / Torula geotricha / Trichosporon matalense |
| Taxonomy: | FUNGI Ascomycota, Saccharomycetes, Saccharomycetales, Dipodascaceae |
| Strain History: | S.P. Abbott (OHS 34) -> UAMH |
| Substrate: | indoor air, RCS strip, from Apis mellifera (honeybee) overwintering facility | Location: | CANADA Alberta, Fairview (GEO: 56.07,-118.392) |
| Isolator: | S.P. Abbott (OHS 34) |
| Isolation Date: | 1993-12-02 |
| Date Received: | 1995-02-23 |
| Characters: | APPLICATION EPA Technology for Mold Identification and Enumeration - // APPLICATION assay name: Geo - reference strain only - Brinkman NE et al, Appl Env Microbiol 69:1775-1782, 2003 // APPLICATION comparison of DNA extraction methodologies for dust samples - Rittenour WR, Park JH, Cox-Ganser JM, Beezhold DH, Green BJ, J Environ Monit 14:766-774, 2012 // APPLICATION QC strain for PCR assay - cited in Patent #6387652 // APPLICATION quantitative PCR analysis of fungi in building materials - Pietarinen V, Rintala H, Hyvärinen A, Lignell U, Kärkkäinen P, and Nevalainen A, Journal of Environmental Monitoring 10:655-663, 2008 // MOLECULAR SYSTEMATICS PCR Method of Identifying and Quantifying Specific Fungi and Bacteria - Haugland RA, Heckman JL, Molecular and Cellular Probes 12: 387-396, 1998 // MOLECULAR SYSTEMATICS PCR Method of Identifying and Quantifying Specific Fungi and Bacteria - Haugland RA, Heckman JL, Wymer LJ, .J. Microbiol. Methods 37:165-176, 1999 // MOLECULAR SYSTEMATICS PCR Method of Identifying and Quantifying Specific Fungi and Bacteria - Haugland RA, Vesper S, Wymer LJ, Molecular and Cellular Probes 13: 332-340, 1999 (Click for publications citing UAMH 7863) |
| Compounds: | |
| Cross Reference: | ATCC MYA-4515 |
| Collections: | Living Strains; Dried Herbarium Material |
| Pathogenic Potential: | Human: no | Animal: no | Plant: no |
| Biosafety Risk Group: | RG1 (check the PHAC ePATHogen Risk Group Database for updates) |
| Regulatory Requirements: | Canadian requesters must provide PHAC Pathogen and Toxin License Number (see: https://www.canada.ca/en/public-health/services/laboratory-biosafety-biosecurity/licensing-program.html) prior to shipment. International requesters must provide all legally required importation documentation prior to shipment. Plant pathogenicity status may be verified by using the USDA Agricultural Research Service (ARS) Fungal Database |
| MycoBank ID: | 182997 |
| Sequences: | >UAMH07863_CS084469_seq85_fr GATATTTCTTGTGAATTGCAGAAGTGA >UAMH07863_CS084470_seq85_fr TTGATTCGAAATTTTAGAAGAGCAAA >UAMH07863_CS084471_seq85_fr CAATTCCAAGAGAGAAACAACGCTCAAACAAG >UAMH07863_AF157596_SSU-LSU CGCCCGTCGCTACTACCGATTGAATGGCTTAGTGAGGCTTCCGGATTGATTAACTGGAGAGGGCAACTTTTCTGGTTGAACGAGAAGCTAGTCAAACTTGGTCATTTAGAGGAAGTAAAAGTCGTAACAAGGTTTCCGTAGGTGAACCTGCGGAAGGATCATTATGAATTANNAATATTTGTGAATTTACCACAGCCAACAACAATCATACAATCAAAACANAAATAATTAAAACTTTTAACAATGGATCTCTTGGTTCTCGTATCGATGAAGAACGCAGCGAAACGCGATATTTCTTGTGAATTGCAGAAGTGAATCATCAGTTTTTGAACGCACATTGCACTTTGGGGTATCCCCCAAAGTATACTTGTTTGAGCGTTGTTTCTCTCTTGGAATTGCTTTGCTCTTCTAAAATTTCGAATCAAATTCGTTTGAAAAACAACACTATTCAACCTCAGATCAAGTAGGATTACCCGCTGAACTTAAGCATATCAATAAGCGGAGGAAAAGAAACCAACAGGGATTGCCTT |
IMAGES:


